Abstract
Background: Mantle Cell Lymphoma (MCL) accounts for 3-10% of all non-Hodgkin lymphomas with a median overall survival of 3-4 years. Hyper-CVAD (CVAD) with or without Rituximab constitutes first line therapy for treatment of MCL, yet the use of this combination is associated with high toxicity and only modest efficacy. On the other hand, impressive clinical efficacy has been reported in relapsed MCL patients treated with rituximab and cladribine (RC). Prediction of response based on cancer genomics heterogeneity creates an opportunity to personalize treatment and avoid toxic therapy which has little chance of response. We conducted a study using the Cellworks Biosimulation Platform to identify novel genomic biomarkers associated with response to CVAD and RC among MCL patients.
Method: Newly-diagnosed MCL patients were selected for this study based largely on genomic data (i.e. aberrations and copy number variations) published in PubMed and TCGA. The Cellworks Computational Omics Biology Model (CBM) is a computational multi-omic biology software model created using artificial intelligence heuristics and literature sourced from PubMed, to generate a patient-specific protein network map. Genomic data from each patient served as input for the CBM. Biomarkers unique to each patient were identified within protein network-maps. Drug impact on the disease network was biosimulated using the Cellworks Biosimulation Platform to determine a treatment efficacy value by measuring the treatment effects on the cell growth score, a composite of cell proliferation, viability, apoptosis, metastasis, DNA damage and other cancer hallmarks. The mechanism of action of each drug was mapped to each patient's CBM and the predicted biological consequences were used to determine response. Biosimulation of CVAD was applied to the patients in this cohort. RC was biosimulated on all CVAD non-responders.
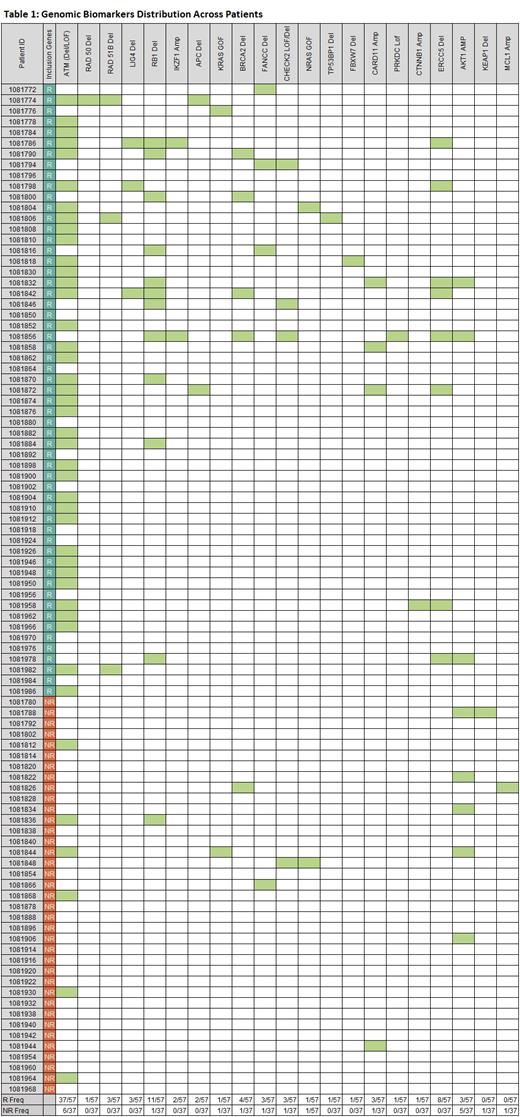
Results: Among the 94 MCL patients treated with CVAD, the Cellworks Biosimulation Platform identified novel biomarkers (Table 1) to predict treatment response or failure. The biosimulation also identified unique drug combinations for patients that were non-responders (NR) to both treatments. Of the 94 patients, 57 were deemed responders (R) and 37 non-responders (NR). ATM LOF/del, RAD51 del, LIG4A del, RB1 del, ERCC5 del, CARD11 amp, IKZF1 amp, and FANCC del were major predictors of CVAD response. These genes contributed to drug efficacy by impacting various pathways, including DNA repair, oxidative-stress, NFKB activation, spindle formation and mitotic-catastrophe. The frequency of aberration affecting these genes was high among the R group and was low in the NR group.
Biosimulation was used to assess response to RC, and predicted, that 41 of 94 patients would respond and 53 would not respond. KMT2D LOF and SMAD4 del were associated with response to RC. Epigenetic dysregulation caused by KMT2D LOF decreased MSH6-mediated mismatch repair required for futile DNA repair leading to replication fork arrest and apoptosis. Interestingly, KMT2D LOF was identified in 20/41 R to RC. MYC amp, NOTCH 1 GOF, and NT5C2 amp were identified as key non-response markers for RC.
In considering both regimens, 27 patients were predicted R to both CVAD and RC, 14 to RC but not to CVAD, 30 to CVAD but not RC, and 23 NR to both regimens. In the latter group, biosimulation predicted that a venetoclax-based combination would be effective in many cases due to the high incidence of TP53 GOF mutation within this subgroup.
Conclusions: This pilot study highlights how the Cellworks Biosimulation Platform applied to the patient-specific CBM can identify treatment alternatives for patients with low likelihood of response to standard therapy or who may be ineligible for CVAD because of co-morbidities. RC responsiveness was either an equivalent but much less toxic option to CVAD or superior to CVAD. By using novel biomarkers derived from comprehensive mutational and copy number analysis, the CBM identified pathway-based, polygenic biomarkers that can be employed to determine optimal drug combinations for MCL patients. This biosimulation approach warrants prospective validation in a larger patient cohort.
Marcucci: Abbvie: Other: Speaker and advisory scientific board meetings; Agios: Other: Speaker and advisory scientific board meetings; Novartis: Other: Speaker and advisory scientific board meetings. Kumar: Cellworks Group Inc.: Current Employment. Castro: Bugworks: Consultancy; Caris Life Sciences Inc.: Consultancy; Omicure Inc: Consultancy; Guardant Health Inc.: Speakers Bureau; Cellworks Group Inc.: Current Employment; Exact sciences Inc.: Consultancy. Khandelwal: Cellworks Group Inc.: Current Employment. Mohapatra: Cellworks Group Inc.: Current Employment. Kapoor: Cellworks Group Inc.: Current Employment. Agrawal: Cellworks Group Inc.: Current Employment. Sauban: Cellworks Group Inc.: Current Employment. Basu: Cellworks Group Inc.: Current Employment. Shyamasundar: Cellworks Group Inc.: Current Employment. Lala: Cellworks Group Inc.: Current Employment. Raju: Cellworks Group Inc.: Current Employment. Palaniyeppa: Cellworks Group Inc.: Current Employment. Ullal: Cellworks Group Inc.: Current Employment. Joseph: Cellworks Group Inc.: Current Employment. Behura: Cellworks Group Inc.: Current Employment. Sahu: Cellworks Group Inc.: Current Employment. Prakash: Cellworks Group Inc.: Current Employment. Mitra: Cellworks Group Inc.: Current Employment. Balla: Cellworks Group Inc.: Current Employment. Patil: Cellworks Group Inc.: Current Employment. Mohanty: Cellworks Group Inc.: Current Employment. Patel: Cellworks Group Inc.: Current Employment. Macpherson: Cellworks Group Inc.: Current Employment. Howard: Sanofi: Consultancy, Other: Speaker fees; Servier: Consultancy; Cellworks Group Inc.: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal